A very basic pipeline of processing EEG data Offline, using MNE-Python.
%matplotlib qt
import mne
import numpy as np
import os.path as op
from matplotlib import pyplot as plt
from mne import Epochs, pick_types, events_from_annotations
from mne.preprocessing import ICA
from mne.preprocessing import create_eog_epochs, create_ecg_epochs
from mne.channels import read_layout
from mne.io import concatenate_raws, read_raw_edf
from mne.decoding import CSP
from sklearn.pipeline import Pipeline
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.model_selection import ShuffleSplit, cross_val_score
print(__doc__)
Automatically created module for IPython interactive environment
# Data Folder Path
data_path = 'EEG_data'
# Specific File Name Appending
fname = data_path + '\AC32-000003-7s-handmov-4ch.vhdr'
raw = mne.io.read_raw_brainvision(fname,preload=True)
Extracting parameters from EEG_data\AC32-000003-7s-handmov-4ch.vhdr...
Setting channel info structure...
Reading 0 ... 197649 = 0.000 ... 395.298 secs...
raw.info
<Info | 16 non-empty fields
bads : list | 0 items
ch_names : list | Fp1, Fz, F3, F7, FT9, FC5, FC1, C3, T7, ...
chs : list | 31 items (EEG: 31)
comps : list | 0 items
custom_ref_applied : bool | False
dev_head_t : Transform | 3 items
events : list | 0 items
highpass : float | 0.0 Hz
hpi_meas : list | 0 items
hpi_results : list | 0 items
lowpass : float | 140.0 Hz
meas_date : tuple | 2019-06-14 15:33:55 GMT
nchan : int | 31
proc_history : list | 0 items
projs : list | 0 items
sfreq : float | 500.0 Hz
acq_pars : NoneType
acq_stim : NoneType
ctf_head_t : NoneType
description : NoneType
dev_ctf_t : NoneType
dig : NoneType
experimenter : NoneType
file_id : NoneType
gantry_angle : NoneType
hpi_subsystem : NoneType
kit_system_id : NoneType
line_freq : NoneType
meas_id : NoneType
proj_id : NoneType
proj_name : NoneType
subject_info : NoneType
xplotter_layout : NoneType
>
montage = mne.channels.read_montage('standard_1020')
print(montage)
<Montage | standard_1020 - 97 channels: LPA, RPA, Nz ...>
raw.set_montage(montage,set_dig=True,verbose=True)
<RawBrainVision | AC32-000003-7s-handmov-4ch.eeg, n_channels x n_times : 31 x 197650 (395.3 sec), ~46.8 MB, data loaded>
raw.plot_sensors()
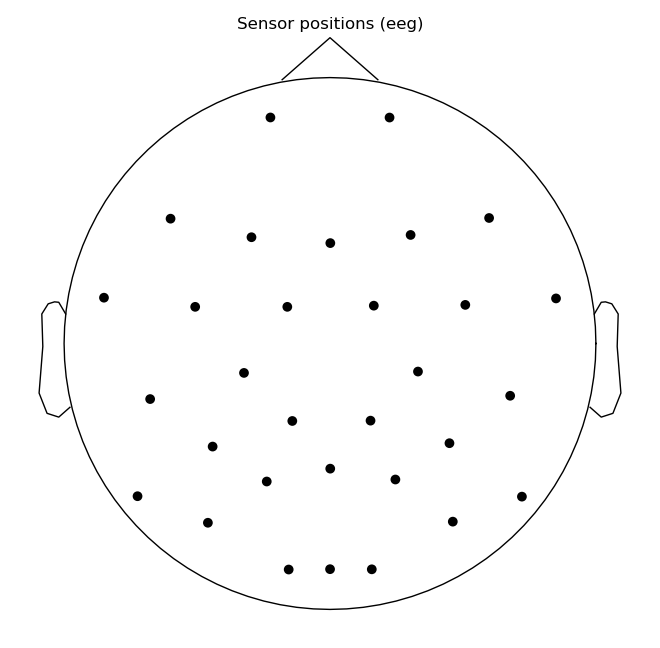
raw.ch_names
['Fp1',
'Fz',
'F3',
'F7',
'FT9',
'FC5',
'FC1',
'C3',
'T7',
'TP9',
'CP5',
'CP1',
'Pz',
'P3',
'P7',
'O1',
'Oz',
'O2',
'P4',
'P8',
'TP10',
'CP6',
'CP2',
'C4',
'T8',
'FT10',
'FC6',
'FC2',
'F4',
'F8',
'Fp2']
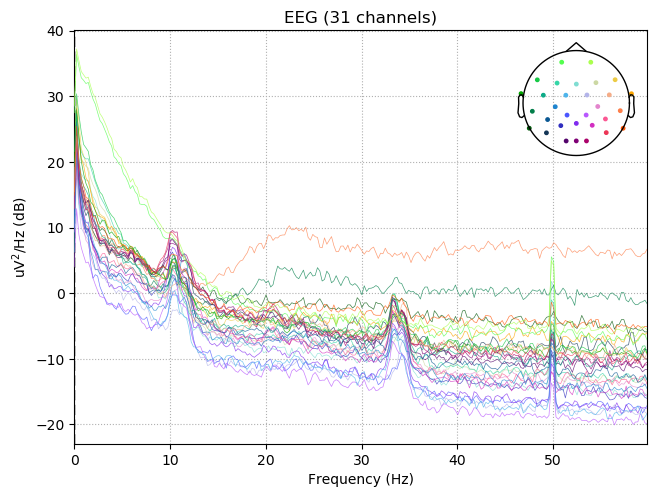
raw.plot_psd(fmax=60)
Effective window size : 4.096 (s)
np.shape(raw._data)
(31, 197650)
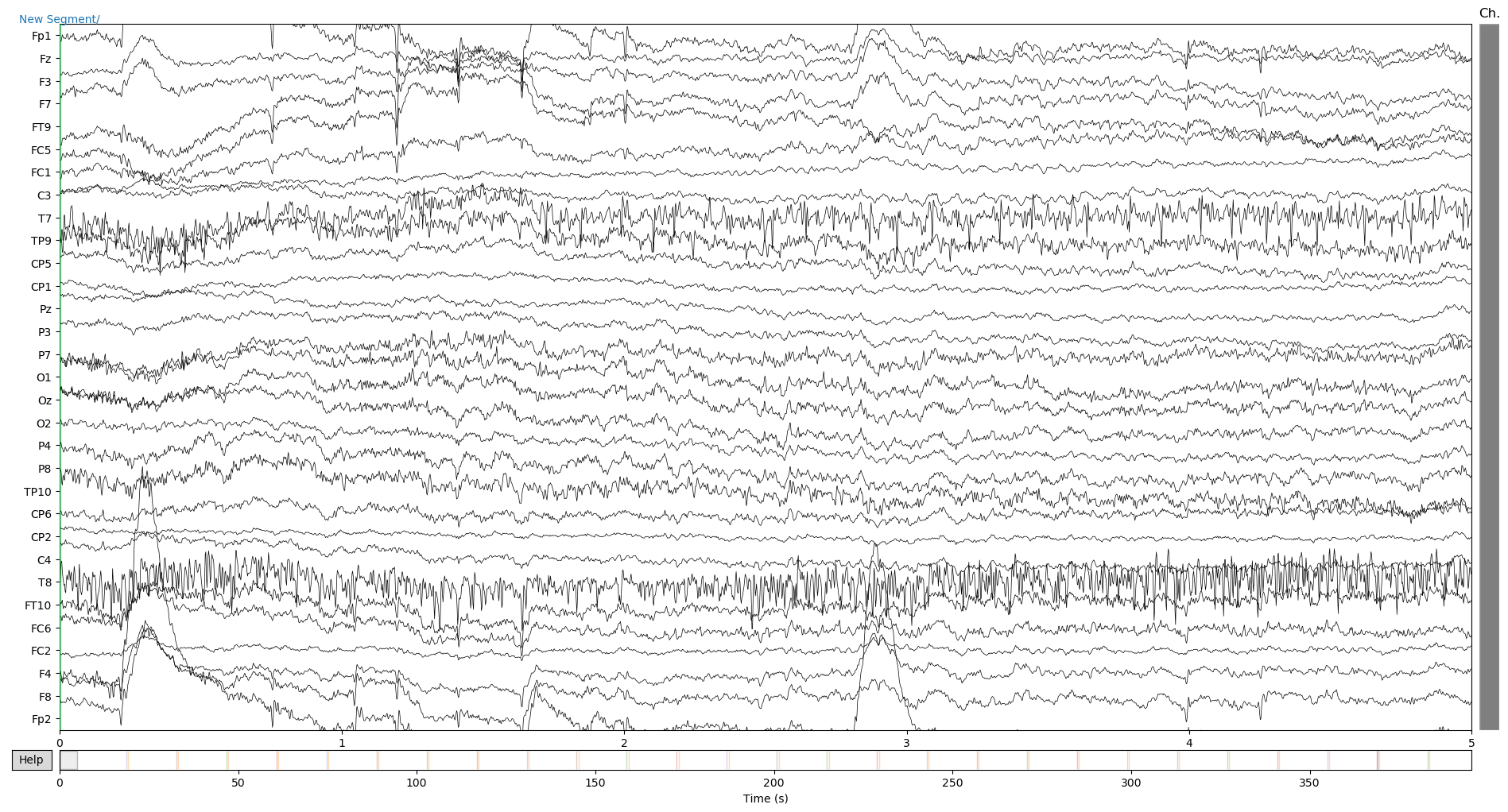
raw.plot(duration=5, n_channels=31)
picks = pick_types(raw.info, meg=False, eeg=True, stim=False, eog=False,
exclude='bads')
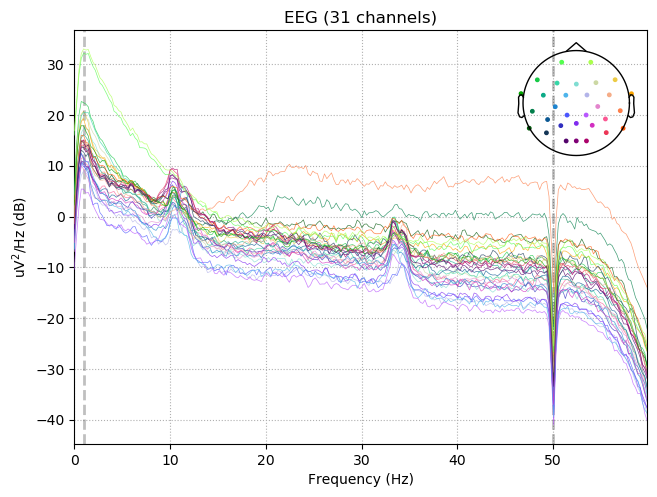
raw.notch_filter(np.arange(50, 201, 50), picks=picks, fir_design='firwin')
raw.filter(1,50., None, n_jobs=1, fir_design='firwin')
raw.plot_psd(tmax=np.inf, fmax=60)
Setting up band-stop filter
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandstop filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower transition bandwidth: 0.50 Hz
- Upper transition bandwidth: 0.50 Hz
- Filter length: 3301 samples (6.602 sec)
Filtering raw data in 1 contiguous segment
Setting up band-pass filter from 1 - 50 Hz
FIR filter parameters
---------------------
Designing a one-pass, zero-phase, non-causal bandpass filter:
- Windowed time-domain design (firwin) method
- Hamming window with 0.0194 passband ripple and 53 dB stopband attenuation
- Lower passband edge: 1.00
- Lower transition bandwidth: 1.00 Hz (-6 dB cutoff frequency: 0.50 Hz)
- Upper passband edge: 50.00 Hz
- Upper transition bandwidth: 12.50 Hz (-6 dB cutoff frequency: 56.25 Hz)
- Filter length: 1651 samples (3.302 sec)
Effective window size : 4.096 (s)
method = 'fastica'
n_components = 25
random_state = 23
ica = ICA(n_components=n_components, method=method, random_state=random_state)
print(ica)
<ICA | no decomposition, fit (fastica): samples, no dimension reduction>
ica.fit(raw)
print(ica)
Fitting ICA to data using 31 channels (please be patient, this may take a while)
Inferring max_pca_components from picks
Selection by number: 25 components
Fitting ICA took 7.0s.
<ICA | raw data decomposition, fit (fastica): 197650 samples, 25 components, channels used: "eeg">
ica.plot_components(inst=raw, psd_args={'fmax': 35.})
[<Figure size 750x700 with 20 Axes>, <Figure size 750x250 with 5 Axes>]
# Include the channels which are Bad (BE CAREFUL not to exclude Real Features in Data useful for classification)
eog_inds = [0,1,2]
ica.exclude.extend(eog_inds)
raw_copy = raw.copy()
ica.apply(raw_copy)
Transforming to ICA space (25 components)
Zeroing out 3 ICA components
<RawBrainVision | AC32-000003-7s-handmov-4ch.eeg, n_channels x n_times : 31 x 197650 (395.3 sec), ~46.8 MB, data loaded>
raw_copy.plot()
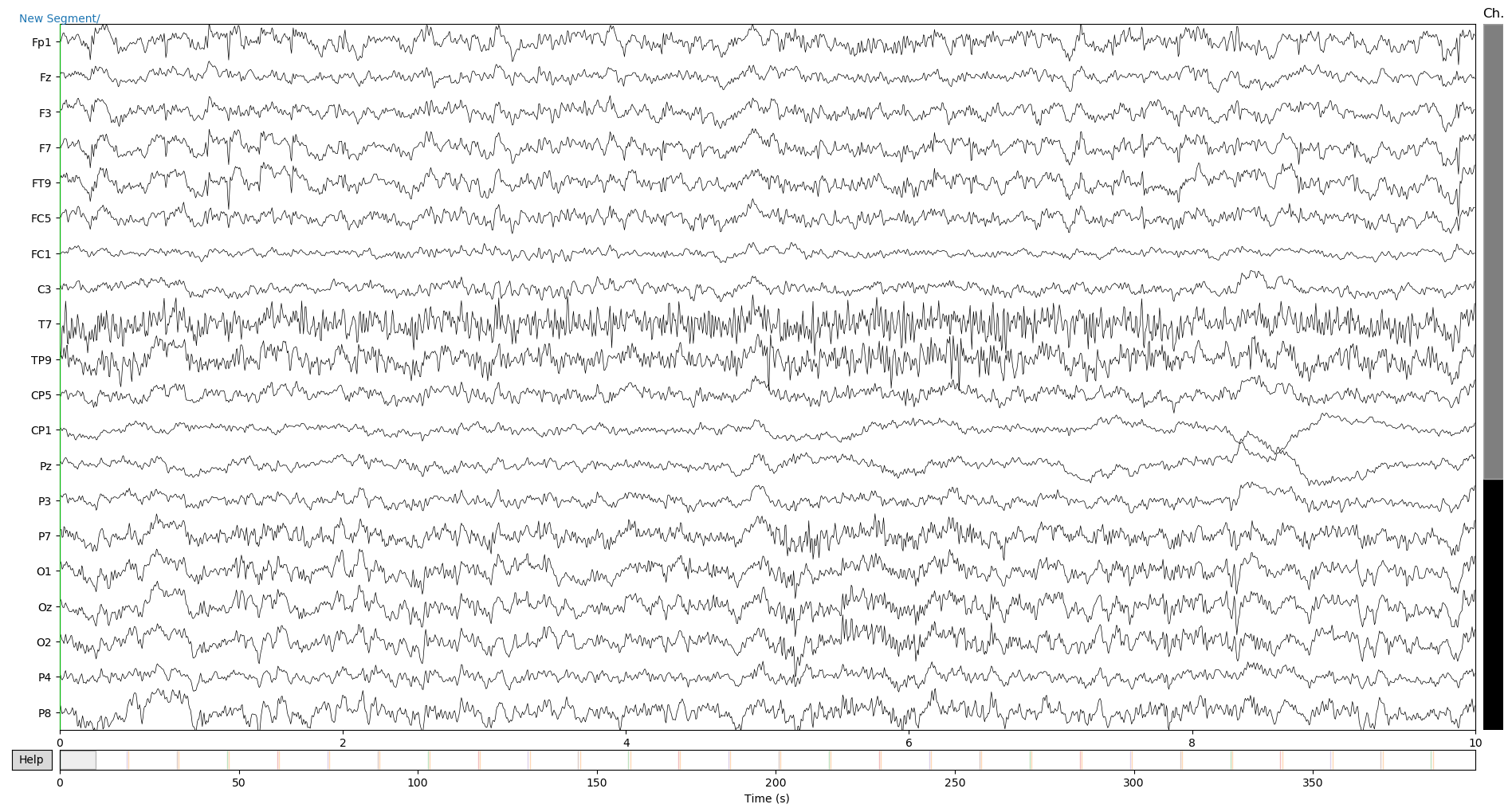
annot = raw.annotations
annot[1]
OrderedDict([('onset', 18.684),
('duration', 0.002),
('description', 'Stimulus/S 20'),
('orig_time', 1560526435.391673)])
annot[7]['onset']-annot[5]['onset']
14.012
raw_copy.annotations
<Annotations | 55 segments : New Segment/ (1), Stimulus/S 20 (7), Stimulus/S 16 (27)..., orig_time : 2019-06-14 15:33:55.391673>
events, _ = mne.events_from_annotations(raw)
Used Annotations descriptions: ['New Segment/', 'Stimulus/S 16', 'Stimulus/S 17', 'Stimulus/S 18', 'Stimulus/S 20', 'Stimulus/S 24']
type(events)
numpy.ndarray
events
array([[ 0, 0, 99999],
[ 9342, 0, 20],
[ 9592, 0, 16],
[ 16356, 0, 24],
[ 16606, 0, 16],
[ 23357, 0, 17],
[ 23607, 0, 16],
[ 30363, 0, 18],
[ 30613, 0, 16],
[ 37378, 0, 20],
[ 37628, 0, 16],
[ 44383, 0, 24],
[ 44634, 0, 16],
[ 51389, 0, 17],
[ 51639, 0, 16],
[ 58389, 0, 18],
[ 58639, 0, 16],
[ 65394, 0, 20],
[ 65644, 0, 16],
[ 72394, 0, 24],
[ 72645, 0, 16],
[ 79394, 0, 17],
[ 79644, 0, 16],
[ 86399, 0, 18],
[ 86650, 0, 16],
[ 93409, 0, 20],
[ 93659, 0, 16],
[100417, 0, 24],
[100667, 0, 16],
[107430, 0, 17],
[107680, 0, 16],
[114434, 0, 18],
[114684, 0, 16],
[121435, 0, 20],
[121685, 0, 16],
[128444, 0, 24],
[128695, 0, 16],
[135452, 0, 17],
[135702, 0, 16],
[142450, 0, 18],
[142700, 0, 16],
[149454, 0, 20],
[149704, 0, 16],
[156455, 0, 24],
[156705, 0, 16],
[163453, 0, 17],
[163703, 0, 16],
[170453, 0, 18],
[170703, 0, 16],
[177455, 0, 20],
[177705, 0, 16],
[184451, 0, 24],
[184701, 0, 16],
[191465, 0, 17],
[191715, 0, 16]])
events_new = events[1::2,:]
# As 4 events are there in sequential manner
events_new[0::4,2] = 1
events_new[1::4,2] = 2
events_new[2::4,2] = 3
events_new[3::4,2] = 4
events_new
array([[ 9342, 0, 1],
[ 16356, 0, 2],
[ 23357, 0, 3],
[ 30363, 0, 4],
[ 37378, 0, 1],
[ 44383, 0, 2],
[ 51389, 0, 3],
[ 58389, 0, 4],
[ 65394, 0, 1],
[ 72394, 0, 2],
[ 79394, 0, 3],
[ 86399, 0, 4],
[ 93409, 0, 1],
[100417, 0, 2],
[107430, 0, 3],
[114434, 0, 4],
[121435, 0, 1],
[128444, 0, 2],
[135452, 0, 3],
[142450, 0, 4],
[149454, 0, 1],
[156455, 0, 2],
[163453, 0, 3],
[170453, 0, 4],
[177455, 0, 1],
[184451, 0, 2],
[191465, 0, 3]])
# Starting the epochs 1 second before stim onset and ending 7 s after onset
# But will crop later to avoid classification of evoked responses by using epochs that start 1s after stim onset
tmin, tmax = -1., 6.
# Giving proper labels to the events
event_id = {'Left': 1, 'Right': 2, 'Up': 3, 'Down': 4}
picks = pick_types(raw_copy.info, meg=False, eeg=True, stim=False, eog=False,
exclude='bads')
epochs = mne.Epochs(raw_copy, events_new, event_id, tmin, tmax, picks=picks,
baseline=None, preload=True)
27 matching events found
No baseline correction applied
Not setting metadata
0 projection items activated
Loading data for 27 events and 3501 original time points ...
0 bad epochs dropped
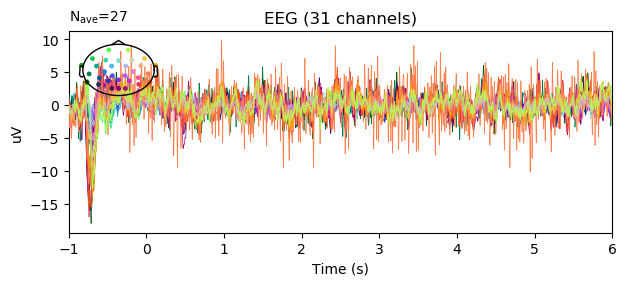
epochs.average().plot(spatial_colors=True, time_unit='s')
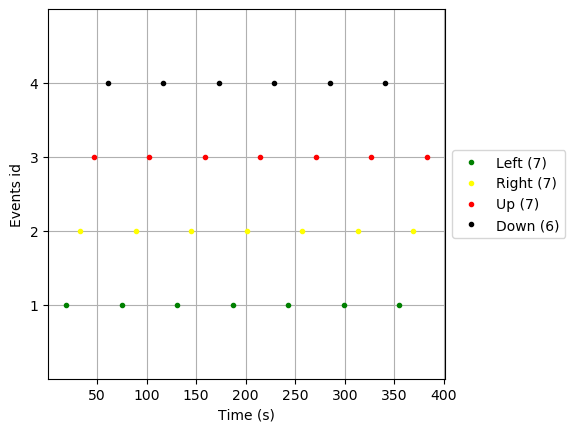
color = {1: 'green', 2: 'yellow', 3: 'red', 4: 'black'}
mne.viz.plot_events(events_new, raw.info['sfreq'], raw.first_samp, color=color,
event_id=event_id)
print(epochs['Left'])
print(epochs.event_id)
print(epochs.events)
<Epochs | 7 events (all good), -1 - 6 sec, baseline off, ~5.9 MB, data loaded,
'Left': 7>
{'Left': 1, 'Right': 2, 'Up': 3, 'Down': 4}
[[ 9342 0 1]
[ 16356 0 2]
[ 23357 0 3]
[ 30363 0 4]
[ 37378 0 1]
[ 44383 0 2]
[ 51389 0 3]
[ 58389 0 4]
[ 65394 0 1]
[ 72394 0 2]
[ 79394 0 3]
[ 86399 0 4]
[ 93409 0 1]
[100417 0 2]
[107430 0 3]
[114434 0 4]
[121435 0 1]
[128444 0 2]
[135452 0 3]
[142450 0 4]
[149454 0 1]
[156455 0 2]
[163453 0 3]
[170453 0 4]
[177455 0 1]
[184451 0 2]
[191465 0 3]]
# Check Visualization here
ev_left = epochs['Left'].average()
ev_right = epochs['Right'].average()
ev_up = epochs['Up'].average()
ev_down = epochs['Down'].average()
f, axs = plt.subplots(2 , 2, figsize=(10, 5))
_ = f.suptitle('Left Right Up Down Hand Movement Imagine', fontsize=20)
_ = ev_left.plot(axes=axs[0, 0], show=False, time_unit='s')
_ = ev_right.plot(axes=axs[0 ,1], show=False, time_unit='s')
_ = ev_up.plot(axes=axs[1, 0], show=False, time_unit='s')
_ = ev_down.plot(axes=axs[1, 1], show=False, time_unit='s')
plt.tight_layout()
epoch3d = epochs.get_data()
np.shape(epoch3d)
(27, 31, 3501)
epochs.plot_image()
27 matching events found
No baseline correction applied
Not setting metadata
0 projection items activated
0 bad epochs dropped
[<Figure size 640x480 with 3 Axes>]
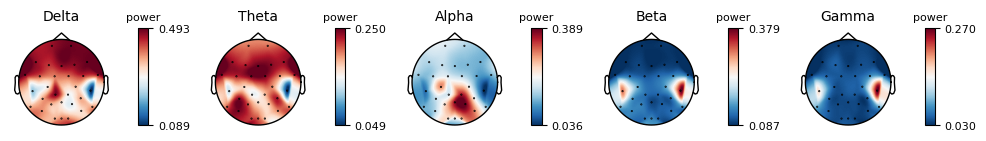
epochs.plot_psd_topomap( normalize=True)
Using multitaper spectrum estimation with 7 DPSS windows
from mne.time_frequency import tfr_morlet, psd_multitaper
l_epochs = epochs['Left']
r_epochs = epochs['Right']
u_epochs = epochs['Up']
d_epochs = epochs['Down']
freqs = np.logspace(*np.log10([6, 35]), num=8)
n_cycles = freqs / 2. # different number of cycle per frequency
power,itc = tfr_morlet(epochs, freqs=freqs, n_cycles=n_cycles, use_fft=True,
return_itc=True, n_jobs=4)
l_power, l_itc = tfr_morlet(l_epochs, freqs=freqs, n_cycles=n_cycles, use_fft=True,
return_itc=True, n_jobs=4)
r_power, r_itc = tfr_morlet(r_epochs, freqs=freqs, n_cycles=n_cycles, use_fft=True,
return_itc=True, n_jobs=4)
u_power, u_itc = tfr_morlet(u_epochs, freqs=freqs, n_cycles=n_cycles, use_fft=True,
return_itc=True, n_jobs=4)
d_power, d_itc = tfr_morlet(d_epochs, freqs=freqs, n_cycles=n_cycles, use_fft=True,
return_itc=True, n_jobs=4)
[Parallel(n_jobs=4)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=4)]: Done 14 tasks | elapsed: 0.8s
[Parallel(n_jobs=4)]: Done 31 out of 31 | elapsed: 1.3s finished
[Parallel(n_jobs=4)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=4)]: Done 7 out of 31 | elapsed: 0.0s remaining: 0.2s
[Parallel(n_jobs=4)]: Done 31 out of 31 | elapsed: 0.3s finished
[Parallel(n_jobs=4)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=4)]: Done 7 out of 31 | elapsed: 0.0s remaining: 0.2s
[Parallel(n_jobs=4)]: Done 31 out of 31 | elapsed: 0.4s finished
[Parallel(n_jobs=4)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=4)]: Done 7 out of 31 | elapsed: 0.0s remaining: 0.3s
[Parallel(n_jobs=4)]: Done 31 out of 31 | elapsed: 0.4s finished
[Parallel(n_jobs=4)]: Using backend LokyBackend with 4 concurrent workers.
[Parallel(n_jobs=4)]: Done 7 out of 31 | elapsed: 0.0s remaining: 0.3s
[Parallel(n_jobs=4)]: Done 31 out of 31 | elapsed: 0.3s finished
u_power.plot_topo(baseline=(-0.5, 0), mode='logratio', title='Average power')
u_power.plot([26], baseline=(-0.5, 0), mode='logratio', title=power.ch_names[26])
fig, axis = plt.subplots(1, 2, figsize=(7, 4))
u_power.plot_topomap(ch_type='eeg', tmin=0.5, tmax=1.5, fmin=8, fmax=12,
baseline=(-0.5, 0), mode='logratio', axes=axis[0],
title='Down Alpha', show=False)
u_power.plot_topomap(ch_type='eeg', tmin=0.5, tmax=1.5, fmin=13, fmax=25,
baseline=(-0.5, 0), mode='logratio', axes=axis[1],
title='Down Beta', show=False)
mne.viz.tight_layout()
plt.show()
Applying baseline correction (mode: logratio)
Applying baseline correction (mode: logratio)
Applying baseline correction (mode: logratio)
Applying baseline correction (mode: logratio)
power_data = u_power.data
np.shape(power_data)
(31, 8, 3501)
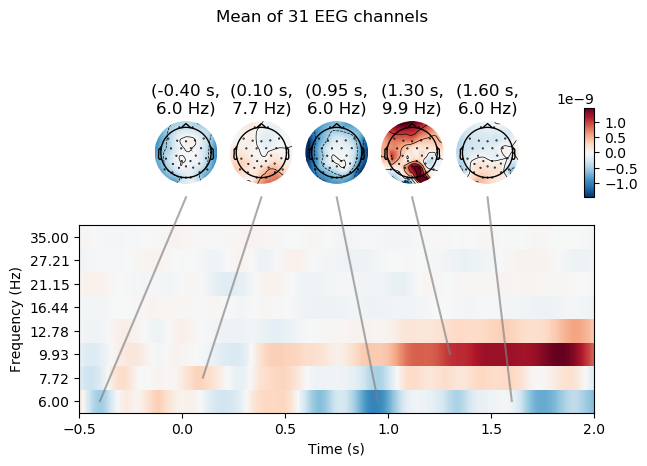
power.plot_joint(baseline=(-0.5, 0), mode='mean', tmin=-.5, tmax=2,
timefreqs=[(.95, 6), (1.3, 9.5), (-0.4, 6), (0.1, 7.7), (1.6, 6.0)])
Applying baseline correction (mode: mean)
Applying baseline correction (mode: mean)
Applying baseline correction (mode: mean)
Applying baseline correction (mode: mean)
Applying baseline correction (mode: mean)
Applying baseline correction (mode: mean)
r_itc.plot_topo(title='Inter-Trial coherence', vmin=0., vmax=1., cmap='Reds')
No baseline correction applied

itc.plot_topo(title='Inter-Trial coherence', vmin=0., vmax=1., cmap='Reds')
No baseline correction applied

raw_copy.save('postica_ayan7shandmov4ch-raw.fif',
overwrite=True)
Writing C:\Users\Nal\Desktop\bciayan\postica_ayan7shandmov4ch-raw.fif
Closing C:\Users\Nal\Desktop\bciayan\postica_ayan7shandmov4ch-raw.fif [done]
epochs.save('epochs-ayan7shandmov4ch-epo.fif',
overwrite=True)